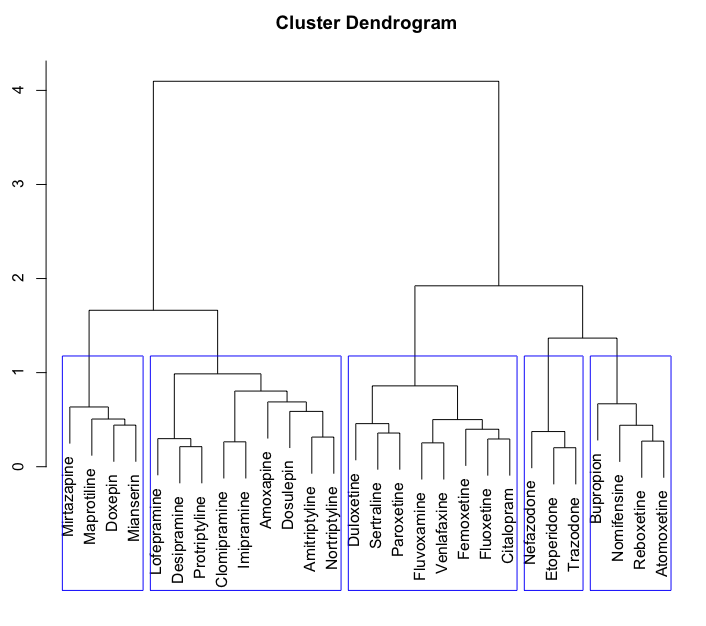
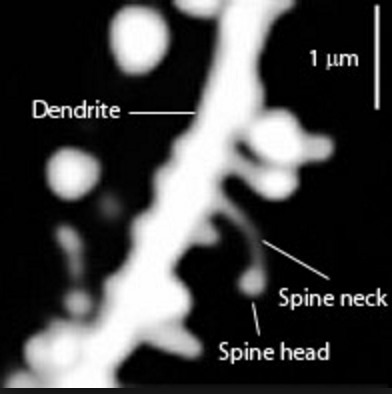
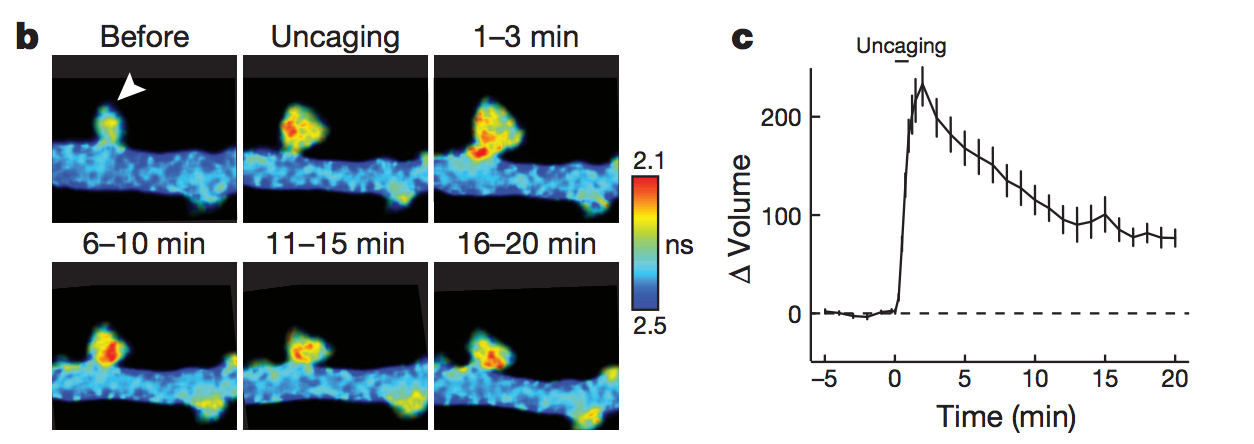
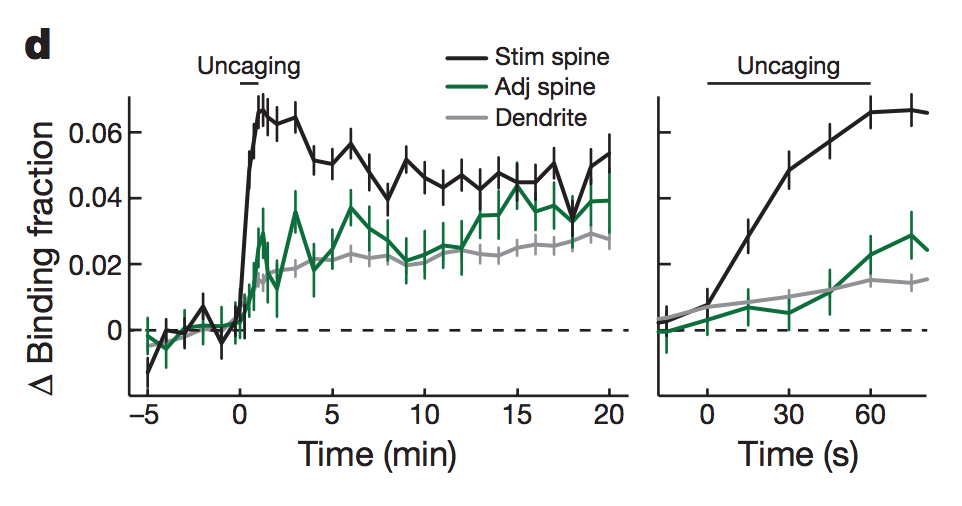
In 2019, Brain Preservation Foundation president Ken Hayworth was tweeting about brain preservation as a potential medical procedure.
Hayworth was asking different scientists who have commented about the topic in the past to engage in a debate on twitter.
I found the discussion between Hayworth and Ken Miller to be especially interesting because it gets into the details of the science and because it is so illustrative of how brain preservation with the goal of potential future revival is discussed. I wanted to document it here to summarize it and for posterity.
It’s hard to capture a non-linear twitter conversation. I did my best. For ease of reading, I’m splitting the conversation into a few different sections.
0. Background: Hayworth’s 2010 article “Why brain preservation followed by mind uploading is a cure for death.”
Amy Harmon’s article about Kim Suozzi and the Brain Preservation Foundation: https://www.nytimes.com/2015/09/13/us/cancer-immortality-cryogenics.html
Miller’s response article: http://www.nytimes.com/2015/10/11/opinion/sunday/will-you-ever-be-able-to-upload-your-brain.html
On June 9th 2019, Hayworth tweeted: “…Still waiting… waiting… for a single neuroscientist to engage publicly in #BrainPreservationDebate. To argue not that it might not work (duh) but to argue why they are so sure it won’t as to withhold the right to choose from terminal patients.”
On June 15th, Hayworth tweeted, “Many of us believe in the long-term success of neuroscience, all the way to mind uploading technology that will eliminate disease/aging. But are clear-minded that this will take centuries. Brain preservation is the ONLY viable bridge for us today. #BrainPreservationDebate”
1. Beginning: On June 16th, Hayworth tweeted to Miller (@kendmil): “@kendmil given your editorial in the NYT https://www.nytimes.com/2015/10/11/opinion/sunday/will-you-ever-be-able-to-upload-your-brain.html … I was wondering if you would be willing to address this question about Aldehyde-Stabilized Cryopreservation’s ability to preserve long-term memories?”
Miller tweeted back: “Sorry but this is beyond my expertise. I don’t know exactly what aldehyde-stabilized cryopreservation does or does not preserve at the molecular level. But I also am doubtful that we know enough to know precisely what must be preserved molecularly to preserve long-term memories.
But even assuming you could preserve, and *enumerate*, every molecular structure/location/state/interaction — I think the bigger question is what would it take to reconstruct a working brain or mind from that. As I argue in that NYT article, we are incredibly far from that.”
2. Timeline: Hayworth responded to the timeline part by agreeing: ”I completely agree. We are probably a century or more away from having the basic neuroscience understanding and technology to scan and simulate a preserved brain. But ASC provides that time and more. That is the argument being put forward for #BrainPreservationDebate”
3. Molecules: Hayworth responded to the molecules part by quoting a recent review: “Thanks for response. ASC preserves everything that glutaraldehyde preserves (connectivity, ultrastructure of synapses, ion channels, mRNA, etc.), it just follows this with inert cryopreservation so brain can be stored for millennia. Seems a wide enough net [to] encompass LTM theories”.
Miller: “What if any disruptions would be expected at the molecular level? Is the idea that it would freeze every molecule in place?? e.g., every CamKII molecule and its phosphorylation state? I also wonder if there could be dynamical interactions that get lost in freezing a snapshot…?”
Hayworth: “Glutaraldehyde (GA) crosslinks proteins in place within seconds and immobilizes other important classes of biomolecules (e.g. mRNA) by trapping them in the fixed matrix. Phosphorylation states appear to be preserved. Quoting from a recent review: “[N]umerous studies have shown that various post-translational modifications are preserved following GA fixation, including phosphorylation (Sasaki et al., 2015)…””
4. Synaptic weight stability: Hayworth also points out: “But you know that CamKII is not in a position to effect millisecond neuronal transmission directly. It is part of feedback loops (http://learnmem.cshlp.org/content/26/5/133.short …) that ultimately stabilize the true functional synaptic weight -dependent on receptor proteins like AMPA.”
To which Matt Krause (@prokraustinator) responds: “Is there really one “true” synaptic weight? I thought they constantly bounce around depending on what you’re recalling from the past, doing now, and planning for the future. If so, W alone isn’t enough; you need dW/dt too. I think this is what @kendmil means by dynamics.”
Hayworth: “That makes no sense from the perspective of storing long-term memories. Weights may change for other reasons (short-term memory) but something has to remain stable to encode long-term memories.”
Krause: “Why not? Stable doesn’t necessarily mean static. Even in computer memory (DRAM), the capacitor voltage is changing all the time (regularly and again when read out), and we’ve designed that to be stable, which isn’t obviously true for biological memories.”
As far as I can tell, Hayworth didn’t respond to this.
5. Molecular correlations: Regarding CamKII feedback loops, Hayworth also argued: “These feedback loops contain a plethora of molecular and structural modifications that all correlate with the functional strength of a synapse. GA would have to erase ALL of this correlated information to prevent the possibility of future decoding.
In fact, there is plenty of evidence that functional synaptic weight is simply correlated with synapse size. https://www.sciencedirect.com/science/article/pii/S0166223603001620 …
https://www.nature.com/articles/nrn2699 …
More recent EM studies: https://cdn.elifesciences.org/articles/10778/elife-10778-v2.pdf …https://science.sciencemag.org/content/360/6395/1349.abstract
“[T]o increase synaptic strength, a synapse must enlarge. The presynaptic terminal enlarges to accommodate more vesicles and active zones. The postsynaptic structure… enlarges to accommodate more receptors, scaffold and regulatory proteins.” https://t.co/mOU4bglW3x [https://mitpress.mit.edu/books/principles-neural-design]
Not sure what “dynamical interactions” you might be referring to that could be required for long-term memory storage? Surgical procedures like https://www.sciencedirect.com/science/article/abs/pii/0013469489900333 shut down neural activity without loss of LTM.
Bottom line: Neuroscience community has already developed really good methods to preserve brains specifically to study the molecular and structural changes involved in learning and LTM. https://www.sciencedirect.com/science/article/pii/S001122401500245X [2015 ASC paper] … allows these to be cryostored indefinitely. #BrainPreservationDebate”
6. Synapses: On June 18th, Hayworth tweeted: “@kendmil Wondering if this adequately addressed your concerns? I am trying to open up a space for calm, rational dialog amongst neuroscientists regarding this. I thought your blanket in statement in the NYT saying brain preservation is impossible today…
“It will almost certainly be a very long time before we can hope to preserve a brain in sufficient detail for sufficient time that some civilization much farther in the future… might have the technological capacity to “upload” that individual’s mind.” [ed: this is Hayworth quoting Miller’s article]
… was misleading and designed to shut down such rational conversation. I am hoping that you might throw me a bone and say you support further dialog within the neuroscience community now? #BrainPreservationDebate”
Miller responded: “At this point I can’t stand behind the statement that it will be a very long time before we can preserve a brain sufficiently, because I don’t feel like I know enough to be certain of that statement. I don’t think it changes any of the main thrust of my article, which was 1/
about how very far in the future is the prospect of being able to reconstruct a mind even from a perfectly preserved brain. I will add, though, you made arguments why you don’t need to perfectly know the status of all the molecules at each synapse, because many factors are 2/
correlated with synaptic strength. But there are two problems with that argument: first, even if all we had to know about synapses was their strength, we have no idea with what precision we would need to know that strength to reconstruct the mind of an individual. Second, we 3/
need to know much more than the strength. As I pointed out in the NYT article, we also need to know how the synapses will learn; and in order to be able to learn quickly while retaining memories for a long time, the synapse appears to need to be quite complex, so that its 4/
internal structure controls how plastic it is and this in turn, along with the synapse’s strength and dynamics, can be controlled by experience. See the work on the cascade model of Fusi and Abbott and more recent work of Benna and Fusi. They have speculated that it is the 5/
need for this complex, dynamic regulation of plasticity as well as of strength that is why the PSD is one of the most complex known biological machines, constructed out of varying numbers of copies of over 1000 different proteins. So it seems quite likely that if you do not 6/
know the full structure and relationships of all of these molecules at the PSD as well as those in the presynaptic terminal, that you would not be able to recreate the brain’s function — the brain would either learn very slowly or forget very quickly. Will your preservation preserve the states and relationships of all of these molecules at every synapse?
Hayworth responded: “Thank you for your thoughtful response. Let me address the three problems you mention:
Q1: “Even if all we had to know about synapses was their strength, we have no idea with what precision we would need to know that strength to reconstruct the mind of an individual.” 1/
A1: Reconstructing “the mind of an individual” to infinite precision is clearly impossible. Our brains are already noisy, chaotic systems. We are continually forgetting old memories and learning new ones and yet we consider our individuality to remain intact. 2/
People willingly undergo brain surgeries like hemispherectomies, to save and improve the quality of their life, with the understanding that some fraction of their personality and memories will change. ‘Success’ in mind uploading should be viewed from this same perspective. /3
A terminal patient choosing brain preservation with the hope of future revival via mind uploading is making the same type of rational judgement –faced with the alternative of oblivion I choose to undergo an uncertain surgical procedure that has some chance of restoring most of /4
the unique memories that I consider to define ‘me’ as an individual. Hopefully this makes clear that I am rejecting a ‘magical’ view of the self. An individual’s mind is computational and, just like with a laptop, an imperfect backup copy is better than complete erasure. /5
Now I believe there is some rough consensus on how perceptual, declarative, procedural, emotional, and sensorimotor memories are stored in the brain and how they interact to give rise to mind (e.g. https://www.sciencedirect.com/science/article/pii/S1074742704000735 …). /6
Such learning and memory is stored as changes to synapses and possibly intrinsic excitability of neurons in recurrent networks which change the attractor dynamics of these networks. /7
Generally, representations in the mind are particular firing patterns of neurons (attractor states) and the process of thought is guided by the attractor dynamics defined by the sum total of the memories laid down over our lifetime. /8
The goal of mind uploading then is to approximate, in a computer simulation of the preserved brain, the attractor dynamics that were present in the original biological brain. /9
We know these attractor dynamics must be relatively robust to noise (e.g. quantal release statistics) and damage (concussion, surgery). /10
These noise considerations imply tremendous redundancy in the encoding of learning and memory, implying that the attractors should be somewhat robust to noise in our determination of the synaptic weight matrix itself. /11
If we wanted to, we could design experiments specifically designed to determine the noise tolerance of brain attractor dynamics to synaptic changes. /12
Measuring the effects of neurotransmitter blockers and optogenetic perturbations on attractor dynamics would be one way of doing so. For example: https://www.nature.com/articles/s41586-019-0919-7 … and https://science.sciencemag.org/content/353/6300/691 … /13
We could also ask whether the signatures of learning and memory can be gleaned from a small sampling of connectivity and synaptic sizes. The answer is yes, again suggesting significant redundancy: https://science.sciencemag.org/content/360/6395/1349… and https://science.sciencemag.org/content/360/6387/430 … /14
Finally, there is some recent evidence that synapse strength is quite tightly correlated with ultrastructural features: https://cdn.elifesciences.org/articles/10778/elife-10778-v2.pdf … /15
Q2: “We also need to know how the synapses will learn; and in order to be able to learn quickly while retaining memories for a long time, the synapse appears to need to be quite complex… work of Benna and Fusi…” /16
A2: There are two points here. First, that synapses are quite complex and that this complexity needs to be modeled accurately in a mind upload or learning will not work. I agree completely, but such complexity can be determined by side experiments on other brains. /17
Second, there may be ‘hidden variables’ besides the synaptic strength that encode information in every individual synapse. This is what Benna and Fusi’s https://www.nature.com/articles/nn.4401 … cascade model says. /18
Their model does not specify how these ‘hidden variables’ are stored but from the things that they do suggest I believe that Aldehyde-Stabilized Cryopreservation would indeed cover that range. /19
After all, we are talking about protein cascades whose dynamics are already being imaged at the synaptic level: https://www.sciencedirect.com/science/article/pii/S0896627315004821 … /20
That said, even if some of the information stored in such hidden variables was lost, the Benna and Fusi simulations imply that this would not significantly disrupt the attractors stored (the hidden variables simply helped later memories not overwrite earlier ones). /21
Q3: “Will your preservation preserve the states and relationships of all of these molecules at every synapse?” A3: A mind upload would model mathematically this complexity to implement the learning rules, but such interactions should be the same across different brains. /22
A brain preservation based on glutaraldehyde fixation should preserve the majority of proteins and their states at each individual synapse –sufficient to determine ‘hidden variables’ beyond synaptic strength if necessary. /23
Specific stains (e.g. http://www.jneurosci.org/content/35/14/5792?utm_source=TrendMD&utm_medium=cpc&utm_campaign=JNeurosci_TrendMD_1 …) could be used to tag key proteins to create a ‘molecularly annotated connectome’ that would reveal such hidden variables along with ultrastructure. /24
I want to thank you for the fascinating discussion and great paper references. I hope you will agree that discussing what would be required for brain preservation and mind uploading should not be a taboo topic. /25
In contrast, it is a topic that can be approached with the current tools of experimental and theoretical neuroscience. We won’t be able to get a definitive answer anytime soon, but we should be able to identify key open questions. /end”
7. Optimism. Miller responds: “You make a reasonable point about brain function being robust to noise given synaptic failures and quantal variability. But it is designed to function w/ that noise. It remains unknown, tho, how much noise in specifying synaptic weights and short-term synaptic dynamics 1/
can be tolerated. I’d also say the idea that representations are in general attractors is very far from clear — in a very few cases there is evidence of representational attractors. But that’s not really critical to your argument. Finally, re Benna & Fusi you argue that 2/
losing hidden variables “would not significantly disrupt the attractors stored (the hidden variables simply helped later memories not overwrite earlier ones).” But that’s the point — if later memories overwrite earlier ones then you are not you, your memories disappear 3/
very quickly. If we can’t start from the snapshot of the person’s last state and proceed forward with normal learning and forgetting — if either they can’t learn new things or rapidly forget old ones — then the living functioning learning remembering person is gone. 4/
More generally, I would just say that you make reasonable arguments but to me you appear extremely extremely optimistic. I take very seriously the depth of our ignorance as to how the brain works overall, how we are able to learn new things quickly w/o quickly forgetting old 5/
things, how the many different forms of memories work and values are computed and decisions made and unified perception achieved and unified actions taken and mood and motivation controlled and on and on … the level of ignorance between our taking specification of a bunch 6/
of molecules and connections and neurons and glia and their states and turning that into a functioning living learning motivated decision-making perceiving mind — that level of ignorance I find astronomical and humbling, and my own gut guess — and it can’t be any more than 7/
that — is that we’re talking time scales of 1000 years, or more, rather than 100. And given our extreme ignorance and given that you know you’re going to lose a fair amount of detailed molecular information, though you don’t seem very clear on exactly what, but given that 8/
it seems to me extremely optimistic to think that you will not lose any molecular or other information that would be critical to reconstructing a functioning sense of self. You’re entitled to your optimism, but that is how it looks to me.
And also, a reminder, we’re not just discussing knowing enuf to make a functioning mind, already every bit as daunting as I just described; but to make Judy’s mind as opposed to Linda’s or Sam’s – to capture the individual’s self, which is a whole other level of complexity.”
Hayworth responds: “Responding to https://twitter.com/kendmil/status/1142645144019197952?s=20 … Optimistic –guilty as charged. But this is sounding like a discussion between two physicists in 1900 debating landing on the moon. Both agree equations allow it, but one insisting the other doesn’t realize how very difficult it will be. \1
You make a reasonable point about brain function being robust to noise given synaptic failures and quantal variability. But it is designed to function w/ that noise. It remains unknown, tho, how much noise in specifying synaptic weights and short-term synaptic dynamics 1/
I hear you, it will be insanely difficult. May take 200 years to get first, 1000 to make routine. Aldehyde-Stabilized Cryopreservation can handle those spans as long as humanity decides that each generation will care for all previous in storage till we can all awake together. \2
I agree it might be impossible given the preservation techniques we have today –won’t know without more research. But I have faith humanity will eventually succeed. First in developing a sufficient preservation procedure, and much, much later in developing a revival procedure. \3
My ‘faith’ is based on careful reading of the literature regarding the synaptic encoding of memory… on careful reading of the aldehyde fixation literature… on personal EM of ASC preserved pig brains… and based on years of developing automated connectome mapping instruments. \4
You “take very seriously the depth of our ignorance as to how the brain works overall”. What about all the progress we have made? For example Lisman’s 2015 review of the field: https://www.sciencedirect.com/science/article/pii/S0896627315002561 … \5
And I wonder how you view the deep learning revolution? Is this not evidence that neuroscience is on the right track: https://www.sciencedirect.com/science/article/pii/S0896627317305093 … Networks now learn object recognition, language translation, driving, chess and go, etc. at human levels. \6
https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1003963 … offers evidence that processing in these artificial networks is similar to processing in the biological brain. https://www.frontiersin.org/articles/10.3389/fncom.2017.00024/full … shows that biologically plausible rules can approximate backprop. \7
Connectome scanning is advancing so rapidly that the NIH is now endorsing a whole mouse EM connectome with human as a long-term goal: https://acd.od.nih.gov/documents/reports/06142019BRAINReport.pdf … Ion milling combined with multibeam SEMs is possible route: https://www.biorxiv.org/content/10.1101/563239v1 … \8
And synapse-level molecular annotation is becoming routine: http://www.jneurosci.org/content/35/14/5792?utm_source=TrendMD&utm_medium=cpc&utm_campaign=JNeurosci_TrendMD_1 … , https://science.sciencemag.org/content/363/6424/eaau8302.abstract … \9
Looking at such rapid progress I find it hard to share your pessimism that it will take 1000+ more years for neuroscience to be successful and that today we neuroscientists wallow in “extreme ignorance”. But you are entitled to your opinion. \10
But the real questions are: Can a terminal patient who, like myself, knows enough about neuroscience to understand the speculative and uncertain nature of the endeavor, give informed consent? \11
Should I have the right to choose brain preservation over certain oblivion, or should that right be withheld from me because someone like you believes ‘it might not work’? \12
Should I have the right to a well-researched, high-quality, regulated preservation procedure, performed pre-mortem in hospital, based on the best techniques that neuroscientists have developed to preserve the molecular and structural correlates of memories (like ASC)? \13
Or should the scientific and medical community continue to turn its back on such research, leaving people like me no option but unregulated, ‘back-alley’, post-mortem cryonics? -the only option people like me have today. \14
Public dismisses brain preservation because they dismiss the core of neuroscience –instead believing that the mind is magic soul-stuff. Opinion pieces like your NYT are playing to the public’s incredulity not of mind uploading, but of the principles of neuroscience itself. \15
But within the neuroscience community I suspect your piece fell a bit flat, especially for young neuroscientists who believe theirs might be the generation to finally understand the brain and who are working hard developing new tools and pushing new computational models. \16
I have met many neuroscientists who chose this field because it addresses the deepest puzzle of them all –the computational mind. Who think solving this puzzle will lead humanity to overcome biological limitations through mind uploading. \17
But these neuroscientist do not dare voice this enthusiasm out loud. Why? Because they are afraid people like you will ridicule them in the press for the sin of ‘taking neuroscience too seriously’. \18
Lest you think I am exaggerating, I assure you I have had many private conversations with neuroscience colleagues who agree with me but explain that saying so publicly will hurt their career. \19
And the brilliant young developer of ASC got ripped to shreds in the press by a mob of ‘magic soul-stuff’ believers, and the neuroscientist who were called on to defend him stabbed him in the back instead. https://www.technologyreview.com/s/610743/mit-severs-ties-to-company-promoting-fatal-brain-uploading/ … \20
Bottom line, this is what I am simply asking: Support research and debate within the neuroscience community regarding brain preservation. Do not suppress it through ridicule. \21
And if called by the press to give an expert opinion, don’t play to the mob of ‘magic soul-stuff’ believers who relish every time a neuroscientist says ‘we have learned nothing”. Instead support your field the way biologists did in the face of evolution deniers. \end”
8. Ethicists: Miller responds (second link): “There is the scylla and charybdis of, on the one hand, giving people false hope, having them spend their time and money pursuing an unachievable (at least currently) immortality; on the other, denying people a choice, to choose to be killed before (but presumably close to) 1/
their natural time of death to allow optimal preservation in pursuit of this hope. At this time I believe it is a false hope, and I choose to explain why I believe that so that others can be informed. They will also hear your perspective. As for choice, I believe that the 2/
terminally ill who are suffering should be allowed to take their own life at a time of their own choosing. You could stretch that to allowing the terminally ill to take their own life for a perfusion procedure, I wouldn’t argue strongly against that. But to ask the medical 3/
establishment, hospitals and doctors, to offer this, to sell this as a service, when it is certainly of unknown efficacy and I think of very dubious efficacy — there are a lot of ethical reasons why that shouldn’t happen. Tho, if perfusion, like other euthenasia, were legal 4/
for the terminally ill then presumably doctors could choose to participate. But even that is very dicey ethically, again because of all the issues around offering false hope and methods of, at the very best, unknown efficacy. I’ll leave sorting that out to the ethicists. 5/
For your other arguments, I haven’t ridiculed anyone, and I think neuroscientists should feel free to express informed opinions on these issues, but they should also welcome debate including different views like mine. Expressing my views is not playing to the mob or 6/
suppressing a field or ridiculing anything. It is precisely the debate you say that you want. I haven’t expressed any opinion on basic animal research on brain preservation. My concern is offering what I believe is false hope to people facing their imminent mortality. You are 7/
free to argue that the hope is not false. Re the other arguments you make, of course we’ve made tremendous progress, and continue to. But that doesn’t much change how fundamentally ignorant we are of how the damn thing works, of what it would take to build a working one. 8/
Progress is rapid but the project is vast. Progress in neural networks is exciting and certainly is suggestive that significant chunks of our intelligence can be understood from distributed, non-symbolic computation. But ask any leader in the field, we are extremely far from 9/
AGI, artificial general intelligence. Though NN’s currently provide our best models of some sensory systems, they are a long ways from the real sensory systems that incorporate top-down as well as bottom-up processing and that learn quickly, constantly and from few examples. 10/
NN’s are exciting but there’s nothing in current NN’s that promises quick progress in understanding the brain.”
Hayworth responds: “Agree, this is an issue for medical ethicists. First step is locking down what is known and unknown on the neuroscience side, then address ethics. Euthanasia rules should apply and all people should have access with no cost. A single philanthropist could ensure this for 1000s.
Fantastic. I hope my reluctant colleagues will take heart that open, civil debate is possible while still pursuing a career.
Agree, glass is half empty/full. But NN are suggestive that complex learned functionality can be encoded in a ‘connectome’, and these systems can be used to explore what fidelity of weights etc. would be necessary for decoding.”
9. Cryonics: Roko Mijic also asks Miller: “”At this time I believe it is a false hope”
What does this really mean?
I don’t like imprecise statements in these kind of debates, because it leaves room for later weaseling by exploiting the vagueness.
Are you genuinely 99+% confident that cryo is impossible? If so say so
Otherwise I think there is a risk that a lot of people hear things like “false hope” and understand it to mean that cryo is totally impossible and unscientific, akin to homeopathy.
But then if it is shown to work one day, “false hope” will be “reinterpreted” to mean that we couldn’t be sure how it would work; a lot of people will have been erased from existence on the basis of some weasel words.
Miller responds (second link): “I’m 99+% confident that no one being cryo’d today will ever be revived. But what good does my saying that do you? @KennethHayworth might say the opposite. So you need to consider arguments, not declarations of conf. For one set of args, see my op-ed https://www.nytimes.com/2015/10/11/opinion/sunday/will-you-ever-be-able-to-upload-your-brain.html … 1/
More generally, I would say: we have no idea what information would be needed to reconstruct a functioning mind from a molecular/cellular brain snapshot. We do know that the problem involves layers and layers of cellular/molecular function that cannot just be reduced to, say, 2/
synaptic strengths and ion channel densities. We also seem unclear on exactly what molecular info is preserved by best cryo techniques. What are odds that that uncertain preservation just happens to capture all of the unknown info needed?
Mijic: “What are odds that that uncertain preservation just happens to capture all of the unknown info needed?”
50%?
Why would you deviate strongly from this in either direction if you don’t know what is needed or what is preserved?”
Miller: “Call it Murphy’s Law, whatever can go wrong, will go wrong (w/ p approaching 1, not .5). Or, if you don’t know what you’re doing, it’s p->1 that you won’t get everything right. Or say they’re N factors you have to get, p=.5 for each, so p=1/2^N of getting them all.”
Mijic: “But what if there is really only one thing you need to know, and N different structures each record that thing, so if you preserve structures at random then the probability of success is 1-2^(-n)
This is the “thinking like a scientist/thinking like a cryptographer” take on it”
Parrish: “This is a good nutshell, and may explain why computer scientists are disproportionately bullish on cryonics compared to other scientists. We expect redundancy to be hard to defeat, and only 1 copy to be needed. Information is leaky….”
Hayworth responds: “Let me clarify: The cryonics community hates me because I challenged them to publish evidence of connectome preservation and they failed. I do not disagree with @kendmil regarding the slim chance of cryonics today. But I think we neuroscientists know how to preserve brains right”
Mijic: “hang on I’m a bit out of the loop: didn’t someone actually win the prize? http://www.brainpreservation.org/large-mammal-announcement/ …”
Luke Parrish: “Look closely, that was ASC — chemically fixed, *then* vitrified. Not compatible with cryonics as-practiced.”
Hayworth also responds: “Q: Do we know enough about the neuromuscular junction to ‘upload’ one? A1: Oh no, it has almost unfathomable molecular complexity. Thousands of journal articles have only begun to scratch the surface.
A2: Of course, it is just a switch. (I support this answer)”
10. Information: Mijic: “4/ Thus I feel that the article doesn’t really engage with the strongest argument for cryonics. Almost everything you talk about is going to be well understood by revival time so it’s irrelevant how complicated it is. What matters is correlation and information.”
Miller: “What I’m talking about in the article is not just general brain function, but the particularities of one brain vs another — not only how strong is each synapse and what are its synaptic dynamics, but what complex state are they in that controls how they will evolve under 1/
further experience. The individual’s brain has to learn not only memories but the structures that keep them stable while also allowing new learning. Similarly, what controls how the excitability of a given cell, or dendrite, will evolve under experience? In other words, the 2/
individual’s brain carries information not only about the strength/excitability of each element but about their mutability, each individually learned. If you can’t reconstruct all of that the brain isn’t going to work correctly. 3/
Your comment about 1 million years and how far we’ve come in 10,000 — we’ve come enormously far in understanding the physical world but we’re a lot less advanced at complex systems, and the brain is likely the most complex of all by far. Do you think we will figure out 4/
*everything* in 800 or 1000 or 10,000 or 100,000 years, or will there always be new scientific frontiers to understand. If the latter — how far down that road is understanding the brain. You just have to understand the enormous complexity, down to cellular/molecular 5/
operations controlling mutability up through the enormous complexity of the neurons and synapses and their short-term dynamics and connectivity and anatomy and on and on. It is easy to underestimate just how deep this complexity is. If you don’t underestimate it, then you come 6/
to believe that the chances of our capturing everything we would need to know to reconstruct an individual mind given our current ignorance are virtually nil.”
Hayworth: “Sorry, let’s stay on the science side. Q: Given a 10nm res EM of mouse retina do you think we could determine whether a given retinal ganglion cell is on center vs off center? …Trying to determine where you think complexity will make impossible.
Miller: “My biggest concern is the amount of information stored intracellularly at the molecular level controlling the dynamics/plasticity of the synapses, dendrites and neurons. So long as you are unsure of how much or which molecular information you can preserve, I think p->1 that 1/
you’ll be missing something essential. That’s as to your preservation method. As to how long it will take until we could take a perfectly preserved brain and make a mind out of it — which requires the ability to reconstruct all the informative bits of the preserved brain as 2/
well as to know how to dynamically assemble them into the individual’s working mind — well, we will get there someday, but I think it is a very very very long time — that it is much deeper and harder than is easy to imagine.”
Hayworth: “We are not really “unsure of how much or which molecular information you can preserve”. Glutaraldehyde preserves proteins, their positions, and their phosphorylation states. This includes ion channels and receptors. It preserves a range of other molecules (e.g. mRNA) in matrix.
It covers all the components suggested to be of importance in just about the full range of existing theoretical models.”
Miller: “Maybe the relevant question is, what wouldn’t it preserve?”
Hayworth: “Changes to protein tertiary structure. Loss of extracellular space. Loss of small ions and molecules. Fixation artifacts arising from first few seconds of living cells reacting to fix. All of this is in the literature. For example: https://cdn.elifesciences.org/articles/05793/elife-05793-v1.pdf …
Miller: “I don’t understand. Your reference compares chemical fixation to rapid freezing. Is either of these your preferred glutaraldehyde method? The paper doesn’t say anything that I can see about preservation at the molecular level. ??”
Hayworth: “High pressure freezing (HPF) is only possible on tiny pieces of tissue (<1mm) but is considered as close to the living biology as you can study. The paper compares glutaraldehyde fixation to HPF to quantify the artifacts in glutaraldehyde fixation.
Aldehyde-Stabilized Cryopreservation begins with glutaraldehyde perfusion fixation so it has all of its artifacts. Follows with a slow perfusion of inert cryoprotectant to allow for long-term storage. Result is basically the same as glutaraldehyde alone but can last indefinitely”.
11. Predicting physiology: Miller: “Another problem with the spine anatomy->physiology hypothesis is that a spine doesn’t have just one “amplitude of its synaptic potential” — it’s dynamic, depressing, facilitating depending on the spike history. The info controlling that is not in the anatomy.”
Hayworth: “There is no evidence that any of this dynamic behavior encodes long-term memory. Probably all are reset during a concussion for example. You make good points about complexity but if they are not related to long-term memory encoding then they are irrelevant.”
Miller: “The dynamic synaptic behavior in response to trains of spikes absolutely will be involved in every aspect of brain function, because every percept, action, decision, memory storage or retrieval or use, act of learning involves sequences of spikes.”
Hayworth: “Again, you can shut down spiking with cold and the person survives with long-term memories https://www.sciencedirect.com/science/article/abs/pii/0013469489900333 …. If we were not discussing uploading but just long-term memory encoding at a conference you would not be bringing up these examples.”
Miller: “The memory is read out and used by patterns, trains, of spikes. The synapses will have changing strengths depending on their spike history. Take those dynamics away and the read out, use, anything the brain does will be changed, probably by quite a lot. The shutdown and 1/
reactivate example doesn’t speak to this. It presumably (?) says you can reset all your synapses to their “I haven’t seen a spike for a long time” state (or maybe freeze them in some other state) and it still works, but the pt is it works by using its synapses with their dynamics”.
12. More molecules. Konrad Kording (@KordingLab) tweets: “I believe that the exact configuration of proteins matter. Time (and in particular anything that disturbs protein configuration) will this delete.
Hayworth: “The exact quantum state as well? Where are you getting this? We have a literature regarding synaptic function. Which proteins are you talking about in particular? What evidence for hypersensitivity to configuration with no correlated information like PSD size? If you give a precise model I can then address the question of whether glutaraldehyde fixation would preserve it.”
Kording: “As long as psd and synapse size do not well predict epsps/ipsps my model is simply “other molecular stuff”. Get me high quality predictions, ideally with in-vivo conditions and I will revoke my objection and become a fan.”
Hayworth: “First a reminder that glutaraldehyde fixation preserves ion channel and receptor proteins in place. You don’t think EPSCs/IPSCs can be reliably predicted based on these? Do you have a required precision based on some model (e.g. attractor memory)? /1
But addressing the PSD and synapse size question, here are some references:
Glutamate uncaging while recording EPSC (slice and in vivo):
https://www.sciencedirect.com/science/article/pii/S0166223603001620 …
Kording: “Nice paper and r2=.8 is good. Wonder how general it is and how it generalizes across situations. But good evidence.”
Miller: “Notice that it’s normalized per dendrite: x-axis, spine size relative to largest spine on the dendrite; y-axis, current relative to largest current of those spines. If all the differences between dendrites came from diffs in space constants to the soma, then in principle could 1/
calculate absolute currents given full knowledge of the neural anatomy and channel distribution, but that’s *if* — not proven. But the other pt is syn strength is both presyn and postsyn. Glut uncaging measures postsyn component, ie. all post glut receptors are activated. 2/
But presyn component involves how many vesicles are released, and that is (at least one thing that) changes (in the mean) with spike history in synaptic depression and facilitation. Need to know presyn behavior also to know synapse behavior.”
Hayworth: “Reviews https://www.nature.com/articles/nrn2699 … that I have read suggest that presynaptic structural changes (e.g. size of varicosity, # vesicles) correlate with postsynaptic ones. For example: https://cdn.elifesciences.org/articles/10778/elife-10778-v2.pdf …”
Miller: “Pt is, presyn release is dynamic. Synaptic depression can be due to vesicle depletion; facilitation can be due to increased p(release) due to increased [Ca++] in the presyn terminal. Different synapses have diff dynamic properties, these can greatly change synaptic function. 1/
See classic works of Tsodyks and of Abbott on many computational effects of depression/facilitation. Also, Markram in ’97 or so showed plasticity could be presynaptic, e.g. increasing p(release) so that first PSP was stronger but facilitation changed to depression”
Hayworth: “Aren’t these for models of working memory as opposed to the types of long-term memory that would be involved in perception, procedural skills, declarative memories, etc. Long-term is what is important for identity preservation. Am I misunderstanding?”
Miller: “The goal is to reconstruct a functioning brain/mind, not just write down a list of memories, right? If your reconstruction scrambles the synaptic dynamics, then the reconstructed brain is going to have very diff activity patterns and compute very diff things than the orig brain.”
Andrew Hires: A coma, medically-induced or otherwise, must scramble synaptic and circuit dynamics. So does a psychedelic experience. Yet, people’s minds are recognizably the same after.
IMHO, the network graph + general synaptic rules might be sufficiently self reinforcing to recover a mind.”
Miller: “I’m not saying you need to maintain your exact dynamic state. You can reset it. I’m saying the dynamic *operations*, starting from wherever you start from, are a key part of the given brain’s function. Scramble that, and, if it works at all, it’s likely a very different brain.”
Hires: “I’d bet there is sufficient information in EM-resolvable synaptic ultrastructure to predict channel composition & synaptic dynamics to 1st order, given sufficient training data and a LOT more work sampling the synaptic properties of region to region projections. Open question.
With fast enough fixation, you could get reserve pool, readily releasable pool, proportion of docked, fused and recycling vesicles. Surely this has predictive power to synaptic depression and facilitation rates, particularly if same projection has been characterized in slice.”
Miller: “Q as always is how much predictive power can you get and how much do you need? But even if you say you can get syn strengths and dynamics, there’s a host of other issues — e.g., all the synaptic and cellular molecular factors controlling the degree of plasticity of each 1/
synapse, dendrite, neuron. As in models of Fusi/Abbott and Benna/Fusi, synapses probably learn their degree of plasticity, and this synapse-by-synapse learning may be critical to ability to learn new things w/o fast forgetting of old. Plasticity of excitability also likely 2/
involves learning. To what extent do you need to take apart the molecular structure of every synapse, dendrite, cell to recreate a mind? And probably many pieces of the story we haven’t even glimpsed yet (e.g., glymphatic system; new Ahrens zebrafish result on glial coding; …)”
Hires: “I agree much that is required to know we have not glimpsed yet. Q is could we bootstrap future discoveries to infer the needed data with sufficient precision from an optimally preserved brain. It’s a fascinating question and a fun debate for (probably) the rest of our lives.”
13. Individual vs generic: Mijic: “I think you’re wrong here @kendmil The goal of cryonics is only to preserve your memories and personality.
Working out exactly how a functional mind works can be done in “side experiments” in the future.
Including side experiments using your own DNA to create a virtual clone of yourself and study its brain, which would almost certainly tell you all of this stuff about how easily new info can be learned, or really anything about the brain other than its memory
if the future has all your memories they can take your DNA, create a virtual copy of you and expose it to the same events and then go measure these dynamic response properties if it’s necessary.
@kendmil I think we need to keep in mind very clearly what the goal of cryonics is: to preserve the unique information that is lost in death. Other information like general facts about brains or anything else that is basically a function of your DNA doesn’t need to be saved
A consequence of this is that any difference which doesn’t differ between identical twins doesn’t need saving. A lot of the things you’re complaining about (e.g. a brain that rapidly forgets things) fail this test.”
Miller: “You’re assuming that your “memory and personality” are staying and everything else is “general brain function”, not individual-specific. When I talk about learning without rapid forgetting, I’m talking about what is probably an individual-specific set of synaptic states
an individual set of synaptic states determine their individual mutabilities learned thru the same processes by which you learn their synaptic strengths. Similar when I talk about dynamics I mean the at least partially learned 2/
patterns of dynamics of individual synapses. There is a lot more than synaptic strengths that defines an individual.”
Hayworth: “In support of @kendmil , he has very clearly stated a legitimate concern: There may be states in synapses that are hidden to EM but important for long-term memory. It would be better if he could say what these states are so we can see if glut preserves them but point taken.”
Miller: “So much is unknown, so it is impossible to point to what these states are. But expt has shown that synaptic dynamics change under learning (e.g. Markram papers, late ’90’s) and theory shows there is a problem combining quick learning with slow forgetting, for which one 1/
theoretical solution is complex internal synaptic states that control mutability.”
Mijic: “So, these states have no correlation whatsoever with anything that is preserved, they are robust over 50+ years of normal life, concussion, brain death, but they are destroyed in an information-theoretic sense by either the aldehyde or the cold or the cryoprotectant?”
Miller: “We don’t know how these states are coded so we can’t know what is or isn’t preserved. But given how much we don’t know, it is very hard to feel confident that current methods preserve all necessary information.”
14. Dynamics: Miller: “I’m not saying you need to maintain your exact dynamic state. You can reset it. I’m saying the dynamic *operations*, starting from wherever you start from, are a key part of the given brain’s function. Scramble that, and, if it works at all, it’s likely a very different brain.
Of course, they are always changing by learning, within cell-type constraints. But, just like synaptic strengths, the strengths of depression and facilitation presumably have some learning-produced structure. Scrambling either strengths or dynamics likely to greatly alter function”.
Parrish: “Even dynamic things have to be encoded in physical reality somehow. That seems to imply molecular structures, my analogy e.g DNA while it is being replicated. So if glutaraldehyde can fix something like that mid step, good chance it fixes dynamic brain operations too…”
Miller: “Of course it’s physical. The question is how much of the necessary information is preserved and reconstructable. As well as, of course, if/when everything needed is preserved,how many eons will it take us to learn to reconstruct all the necessary info and create a mind out of it.”
Parrish: “Perhaps a relevant data point would be what kinds of things are known not to be preserved by glutaraldehyde. It seems broad-acting on the level of visible detail seen through EM, but are there many important classes of molecule that are not acted upon?”
Hayworth: “Recent review: https://osf.io/8zd4e I love that we have gone from “cryonics can’t be trusted because it can’t demonstrate it preserves what every neuroscientist knows is crucial (synaptic connectivity)” to “ASC can’t be because the neuroscience textbooks may be totally wrong”
I think more neuroscientists should just embrace the fact that people are taking their models seriously. Our minds are just a product of neuronal computations defined by connectivity, ion channels, etc. And neuroscience has figured out how to preserve these indefinitely. Go team!
And team #neuroscience is just getting started. In the coming decades we will figure out how to map a glutaraldehyde-preserved mouse brain at the synaptic level and how to annotate this with whatever molecular info is needed to decode moderately complex learning and memories.
Many decades later team #neuroscience will figure out how to simulate a mouse brain from such a molecularly-annotated connectome. And perhaps by the beginning of next century we will be ready to upload the first human in a $100 billion Apollo-scale project.
That project to “put the first person in cyberspace and return them safely to consciousness” will answer all of our philosophical questions about mind uploading. A century later when uploading becomes routine our ancestors will ask one question…
Why didn’t our ancestors in the early 21st century adopt brain preservation? And they will arrive at one answer: We were not killed by a lack of knowledge or technology, we were killed by our bad philosophy: http://www.brainpreservation.org/wp-content/uploads/2015/08/killed_by_bad_philosophy.pdf …”
15. No copy problem: Michael Hendricks: “You are not the “same” if you can simultaneously exist as different entities…that is a physical impossibility. And there is nothing magically different about whether the sim exists before or after you’re dead.”
Hayworth: “I have to disagree with that. I can have multiple drafts of a program on several computers, some running simultaneously. They are all the ‘same’ when compared to starting over from scratch. I see no reason that same argument does not apply to us.
Q: If we assumed that the philosophical copy problem really did forbid ‘survival by backup copy’, what would this mean for a race of sentient robots where, unlike biology, copying programs and data are trivially easy? Doesn’t the copy problem imply sentient robots cannot exist?”
Kording: “No two robots are *identical*. I have no idea why that would limit their sentience.”
Hayworth: “No one cares about ‘identical’. If I make a backup copy of C3PO’s memories before a mission, he gets destroyed, and then the backup is put into another robot body then 99.9% of what made C3P0 unique is still here to interact with. Same with us right?”
Kording: “I am totally with you. I just had to agree that @MHendr1cks was right about “identical not possible””
Miller: “I agree that there is no copy problem. In the science fic world where we could replicate your brain, it wakes up as you the same way you wake up as you in the morning — the you that went to sleep doesn’t exist anymore, something else is waking up with an experience of being 1/
continuous with the you that went to sleep. That wouldn’t be different if it’s a brain replica in 1000 years or you in the morning. Of course, each copy then goes off to have its own experience and individuates, like identical twins. But for now I think this is sci fic.”
16: Simulations not dispositive: Hayworth: “The ball is no longer with cryonicists or skeptics, it is now in the neuroscientists’ court. Do we believe our research (http://www.brainpreservation.org/quotes-on-synaptic-encoding-of-memory/ …) and believe our field will eventually be successful (http://www.brainpreservation.org/wp-content/uploads/2019/03/aspirationalneuroscienceprize_overviewdocument.pdf …) or not. That is the story that is unfolding today.”
Kording: “Hey. I don’t see why my challenge is not valid. Show in a small system that you can simulate the ephys based on connectomics. I do not argue that cryonics is per principle impossible. I argue that central and easily testable assumptions have not been tested.”
Hayworth: “Please say ASC or something more precise. Saying cryonics is asking for misunderstanding.
People are dying today and want to take chance on ASC. Folks like Nectome are gearing up to answer that demand. Now is the time for the neuroscience community to set clear goal line they must cross.
Your ‘ephys based on connectomics’ challenge may be best, but needs clear success criteria. Must be answering a real concern not just a feel good demo. Again, people are dying now. The ‘preserves synaptic connectivity’ challenge was met and now the goal posts have moved.”
Kording: “And the critique that we know too little to have any confidence that EM structure or even the joint set of everything that is preserved with cryonics is sufficient stands and seems scientifically tenable.”
Miller: “Yes. I don’t think whether or not we can predict ephys from connectomics today is the right test. We can’t, but someday we’ll be able to. To me the real concern is how very little we know about how the brain works, and the likelihood that critical things currently unknown 1/
won’t be preserved. Unfortunately there’s no challenge or test for that, since it’s unknown unknowns. Tho we can point to some likely issues like complex synapse-specific internal states. There are other issues I can see, like how you get info in & out of simulated brain 2/
if you don’t preserve sensory input structures (retina, cochlea) to know which input neurons should carry which info and spinal cord and ganglia to know same for output neurons, but advocates can probably address that; or how many eons before we can read out preserved info 3/
and successfully simulate the operating, learning brain from it and the likelihood that civilization and the preserved brains both last that long, but if you’re optimistic enough that won’t look like a fatal problem. The main issue I think is how much is unknown.”
Kording: “but I think we can agree that noone can remember their own cryopreservation – there is too little time for proteins to be made or for structure to change.”
Hires: “That’s a feature not a bug”
Jprwg: “A question, which I hope makes sense: to what extent does the info cryonics can plausibly capture of a brain’s structure encode how the brain works, vs just encoding that individual? Do we at all get working brains ‘for free’ or will their full design need be explicitly modeled?”
Miller: “Definitely not for free. We need to understand how all the pieces make a dynamically working brain.”
Jprwg: “Thank you. To clarify: we could analogise general brain workings vs individual identity to a piece of software that can load & run different users’ data, eg a word processor. Are you saying then that cryonics gives us only the user data, not any of the software functionality too?”
Miller: “No, software/hardware separation is a bad analogy for the brain. To save enough to recreate an individual, you would presumably have to save everything that makes a working brain”
17. Conclusion: Hayworth: “Seems this thread has drifted from hardcore neuro…less productive. @kendmil would you agree that https://www.biorxiv.org/content/10.1101/556795v1.abstract … demonstrates that function-from-connectome is possible at least to some level of precision?”
Miller: “It’s not in dispute that function derives from structure. Q is, how much and what structure do you need to re-create a dynamically working, learning, particular individual’s mind, and how long to develop the knowledge to read out info and re-create a mind.”
Hayworth: “Yes, and papers like https://www.biorxiv.org/content/10.1101/556795v1.abstract … address this question. If learning was stored as subtle hidden synaptic molecules, or was incomprehensibly complex (as you have been alluding to) then they should have found nothing using primitive rabies virus tracing right?
“Q: what structure do you need” -paper suggests connectome is sufficient for visual RFs. ASC can preserve this (and much more), EM can image and computers can simulate this at small scale today.
I am trying to zero in on core of your objection so I can either address it directly (as I have tried with refs) or succumb to its irrefutability. Each element of your statement “re-create a dynamically working, learning, particular individual’s mind” I am trying to address. \1
The neural models we have today are “dynamical and learn” while based solely on things ASC preserves (morphology, connectivity, synaptic ultrastructure, receptors, ion channels). You countered there may be hidden variables that would prevent function prediction based on these. \2
I countered with studies that showed particular functions (e.g. https://www.biorxiv.org/content/10.1101/556795v1 …) could be determined based on a subset of these. Benna & Fusi is a theoretical ‘what if hidden variables?’ which can be refuted by evidence correct? \3
A “particular individual’s mind” is unique because of learning-related changes (Unless you are implying the philosophical copy problem?). I provided refs (e.g. El-Boustani 2018) showing such learning is encoded in interpretable structural changes preserved by ASC. \4
“how long to develop the knowledge” is being addressed by showing how far we have come already (e.g. understanding the synaptic basis of memory sufficiently to create false ones https://royalsocietypublishing.org/doi/full/10.1098/rstb.2013.0142 … , and to label and erase the synapses encoding one https://www.nature.com/articles/nature15257 …) \5
“how long to…read out and re-create a mind” is addressed by advances in connectomics and molecular imaging that are in principle compatible with ASC preservation. Given our progress in all of these areas does my estimate of ‘one to several centuries’ really seem outlandish? \6
Q: Is evidence of RFs, auditory and contextual fear memories, etc. irrelevant to your objections because you believe that consciousness is built of different circuit elements and molecules than those studied by neuroscientists today? If so I can address that as well. \7
Miller (otherlinks): “Ken, you are missing something basic to what I am saying, so let me try again. You are focused on long-term memories. But what you claim to want is to reconstruct a mind/brain. A mind/brain is a lot more than a fixed set of long-term memories. It has to operate in the world, 1/
creating motivations, making decisions, learning from ongoing experiences while retaining and utilizing and modifying (reconsolidation) prior memories. I don’t have a problem w/ idea that much of memory is in synapses, strengths & dynamical properties. But a mind that proceeds 2/
with those to new experiences and continual learning has to know how to modify synapses to learn from new experiences without losing old memories. We know theoretically there is a big problem with achieving that. One set of solutions are those of Fusi, where each synapse has 3/
learned a complex internal state that among other things codes that synapse’s mutability. If that were true, and you lost that internal state info, either the reconstructed brain could learn little new or it would quickly forget the old. More generally, a synapse is one of 4/
the two most complex molecular machines known — at least, a mammalian synapse — and those 1000+ different proteins, multiple copies of each, must be storing a lot more info than a strength. Even if it were true that the memories are largely in the strengths, the *function* 5/
of the synapse, the way it supports both new learning and memory maintenance/updating in the experiencing organism, must be far more complex than a scalar strength. And the functions involved in a brain operating itself and taking actions in interaction w/ experience – we have 6/
almost no understanding of how these work. Neurons are complex cells with complex internal signaling and changes in gene expression are clearly part of learning, again in ways we only dimly have glimpses of. What I am trying to say is your focus on what stores the info in 7/
long-term memories is missing 99+% of the action in what it would take to reconstruct a functioning mind. And most of that 99+% is yet unknown, so we have no way of saying what is critical to its preservation. 8/
Let me add, a lot of the examples you cite involve circuitry correlating with, being predictive of, receptive field (RF) properties. I’ve developed a number of the models of how circuits create various functional response properties of visual cortical cells. So I know these 9/
issues well. I believe we have learned a lot about how some basic functional response properties of V1 cells are constructed. But the fact is we still are very poor at predicting V1 responses to natural scenes, we only dimly understand how its responses are modulated by 10/
numerous other factors and areas, we have very limited understanding of how attention is achieved, and we haven’t even begun to understand how a unified visual percept is created out of ambiguous and sometimes rivalrous possibilities. In other words, we have lit up a bit of the darkness, but the surrounding darkness is vast, and we don’t even know how vast.”
Hayworth: “We know theoretically there is a big problem” -Sorry, sounds like literature I am unfamiliar with. Could I get an experimental ref? Experimentally determined cap differs by how much? Isn’t hippocampal replay an alternative solution to Fusi?
Miller: “See Fusi & Abbott 2007, Nature Neuro. Fusi, refs 5-7, identified problem w existing memory models given synapses with finite # of levels (bounded, finite resolution) – # memories scales as log(N) instead of N, where N is # of synapses. This paper more closely examines problem. 1/
This log(N) basically gives tradeoff between learning & forgetting. If learn quickly, must forget quickly. Fusi, Drew & Abbott 2005 and Benna & Fusi 2016 use complex internal synaptic states to ameliorate or solve problem.
Anyhow, I don’t really want to spend a lot more time arguing about these things. I would like you to correctly understand what I am saying, but don’t really have more energy for this otherwise.”
Hayworth: “I understand. I very much appreciate you taking the time to explain your objections. I think I understand them and will incorporate them into my thinking going forward. If you are going to be at SfN this year perhaps we can discuss more over a beer. I’ll buy the first round.”